Dust emission¶
Synthesizer can model thermal dust emission using an energy balance approach. This is done using dust emission models, which can then be handed to an EmissionModel to produce a dust emission spectrum, which can be combined with any other spectra in the model.
First we load some common modules, and define a wavelength array over which we will generate our dust emission SED
[1]:
import matplotlib.pyplot as plt
import numpy as np
from unyt import Angstrom, K, Lsun, Msun, um
from synthesizer.emission_models.dust.emission import (
Blackbody,
Casey12,
Greybody,
IR_templates,
)
lam = 10 ** (np.arange(3.0, 8.0, 0.01)) * Angstrom
Parametric models¶
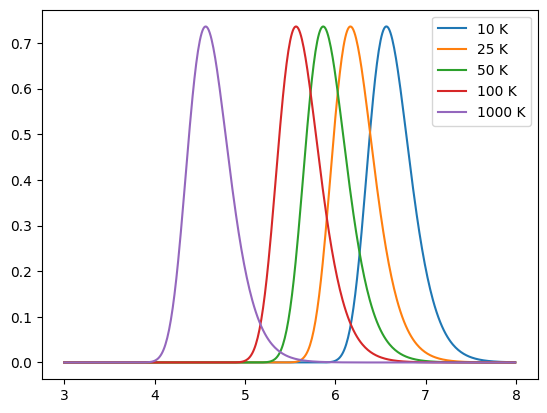
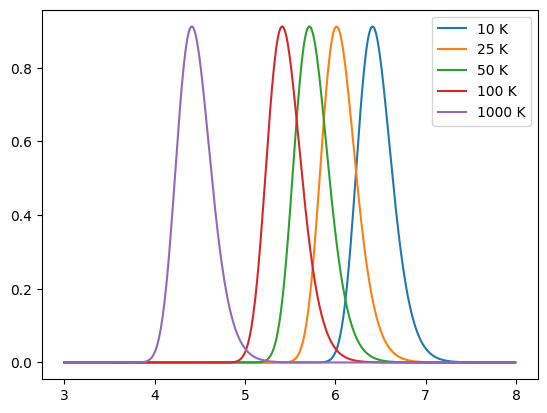
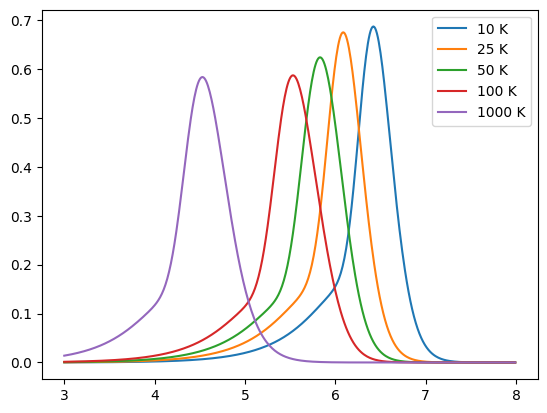
Below we show a number of simple models, including a blackbody, greybody (non-unity emissivity) and the model detailed in Casey et al. (2012). By default a dust emission model provides a normalised spectrum.
[2]:
for T in [10, 25, 50, 100, 1000]:
model = Blackbody(T * K)
sed = model.get_spectra(lam)
plt.plot(np.log10(sed.lam), sed.luminosity, label=f"{T} K")
plt.legend()
plt.show()
[3]:
for T in [10, 25, 50, 100, 1000]:
model = Greybody(T * K, 1.6)
sed = model.get_spectra(lam)
plt.plot(np.log10(sed.lam), sed.luminosity, label=f"{T} K")
plt.legend()
plt.show()
[4]:
for T in [10, 25, 50, 100, 1000]:
model = Casey12(T * K, 1.6, 2.0)
sed = model.get_spectra(lam)
plt.plot(np.log10(sed.lam), sed.luminosity, label=f"{T} K")
plt.legend()
plt.show()
[5]:
[8, 1000] * um
[5]:
unyt_array([ 8, 1000], 'μm')
Adding CMB heating¶
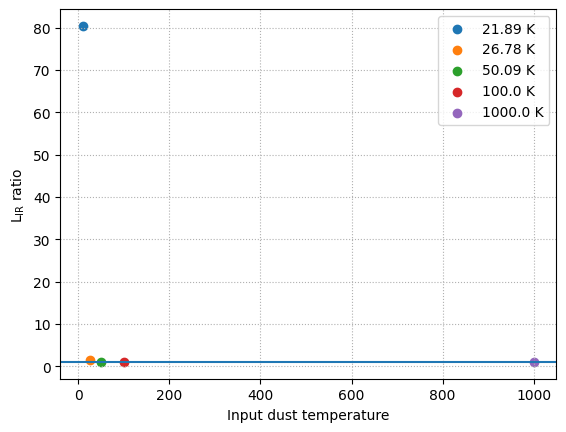
At high redshift, the effect of CMB heating is important. To account for this any dust emission model can incorporate the impact of CMB heating (cmb_heating) at a given redshift (\(z\)). Below we compare the dust emission spectrum from the Casey+12 model with and without CMB heating:
[6]:
for T in [10, 25, 50, 100, 1000]:
model = Casey12(T * K, 1.6, 2.0)
model_cmb = Casey12(T * K, 1.6, 2.0, cmb_heating=True, redshift=7)
sed = model.get_spectra(lam)
sed_cmb = model_cmb.get_spectra(lam)
L_ir_ratio = sed_cmb.measure_window_luminosity(
window=[8, 1000] * um
) / sed.measure_window_luminosity(
window=[8, 1000] * um
) # same as model_cmb.cmb_factor
plt.scatter(
T, L_ir_ratio, label=f"{np.around(model_cmb.temperature_z, 2)}"
)
plt.axhline(y=1)
plt.grid(ls="dotted")
plt.xlabel("Input dust temperature")
plt.ylabel(r"L$_{\rm IR}$ ratio")
plt.legend()
plt.show()
IR Templates¶
Draine & Li 2007 dust models¶
The more complex model present in Draine & Li 2007 is also available; this requires a grid to be loaded in.
Begin by reading in the DL07 grids, which have been created by downloading the ASCII DL07 files and running
from synthesizer.utils import process_dl07_to_hdf5
process_dl07_to_hdf5()
However, you can download these preprocessed grids using the synthesizer-download command line tool.
synthesizer-download -d /path/to/destination --dust-grid
[7]:
from synthesizer.grid import Grid
grid_name = "MW3.1"
grid_dir = "../../../tests/test_grid/"
grid = Grid(grid_name, grid_dir=grid_dir, read_lines=False)
print(grid.axes)
['qpah', 'umin', 'alpha']
Below we show how to use the DL07 model directly, and plot the dust emission spectrum for a number of dust luminosities. You can also pass these models to an EmissionModel and they will be used automatically.
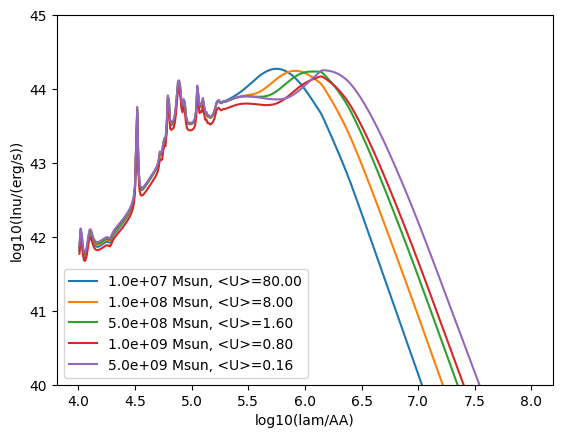
[8]:
for mdust in [1e7, 1e8, 5e8, 1e9, 5e9]:
model = IR_templates(
grid, mdust=mdust * Msun, ldust=1e11 * Lsun, verbose=False
)
sed = model.get_spectra(lam)
plt.plot(
np.log10(sed.lam),
np.log10(sed.luminosity),
label="{:.1e} Msun, <U>={:.2f}".format(mdust, model.u_avg),
)
plt.xlabel("log10(lam/AA)")
plt.ylabel("log10(lnu/(erg/s))")
plt.ylim(40, 45)
plt.legend()
plt.show()