Note
Go to the end to download the full example code.
Plot spectra by age¶
This example plots all the spectra for a single metallicity.
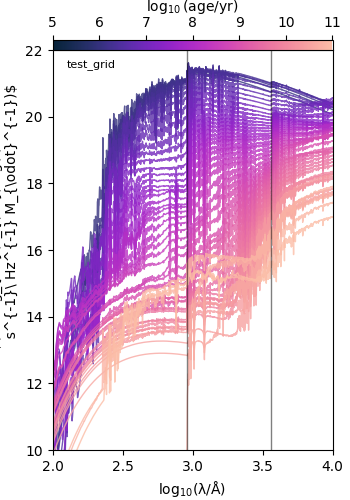
test_grid
target metallicity: 0.01
metallicity: 0.01 dimensionless
import argparse
import glob
import os
import cmasher as cmr
import matplotlib as mpl
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import numpy as np
from synthesizer.grid import Grid
# colourmap to use
cmap = cmr.bubblegum
# mapping of age to colour
norm = mpl.colors.Normalize(vmin=5.0, vmax=11.0)
def plot_spectra_age(grid, target_Z, spec_name="incident"):
# get closest metallicity grid point
grid_point = grid.get_grid_point(
log10ages=grid.log10age[0],
metallicity=target_Z,
)
# metallicity grid point
iZ = grid_point[1]
# get actual metallicity for that grid point and print it
Z = grid.metallicity[iZ]
print(f"target metallicity: {target_Z:.2f}")
print(f"metallicity: {Z:.2f}")
# initialise plot
fig = plt.figure(figsize=(3.5, 5.0))
left = 0.15
height = 0.8
bottom = 0.1
width = 0.8
# define main ax
ax = fig.add_axes((left, bottom, width, height))
# define colourbar ax
cax = fig.add_axes((left, bottom + height, width, 0.02))
# add colourbar
fig.colorbar(
cm.ScalarMappable(norm=norm, cmap=cmap),
cax=cax,
orientation="horizontal",
)
# colourbar formatting and labelling
cax.xaxis.tick_top()
cax.xaxis.set_label_position("top")
cax.set_xlabel(r"$\rm \log_{10}(age/yr)$")
# loop over log10ages
for ia, log10age in enumerate(grid.log10age):
# get spectra
Lnu = grid.spectra[spec_name][ia, iZ, :]
# Lnu = fnu_to_flam(grid.lam, Lnu)
# plot spectra
ax.plot(
np.log10(grid.lam),
np.log10(Lnu),
c=cmap(norm(log10age)),
lw=1,
alpha=0.8,
)
# plot Lyman and Balmer limits for reference
for wv in [912.0, 3646.0]:
ax.axvline(np.log10(wv), c="k", lw=1, alpha=0.5)
# add model name
ax.text(2.1, 21.5, grid.grid_name, fontsize=8)
# set wavelength range (log(Angstrom))
ax.set_xlim([2.0, 4.0])
# set luminosity range
ax.set_ylim([10.0, 22])
# add labels
ax.set_xlabel(r"$\rm log_{10}(\lambda/\AA)$")
ax.set_ylabel(
r"$\rm log_{10}(L_{\nu}/erg\ \
s^{-1}\ Hz^{-1} M_{\odot}^{-1})$"
)
# return figure and axes for further use
return fig, ax
if __name__ == "__main__":
# Get the location of this script, __file__ is the absolute path of this
# script, however we just want to directory
# script_path = os.path.abspath(os.path.dirname(__file__))
# Define the path to the test grid
# test_grid_dir = script_path + "/../../tests/test_grid/"
test_grid_dir = "../../tests/test_grid/"
parser = argparse.ArgumentParser(
description=(
"Create a plot of all spectra for a given metallicity in \
a grid"
)
)
# The name of the grid. Defaults to the test grid.
parser.add_argument(
"-grid_name",
"--grid_name",
type=str,
required=False,
default="test_grid",
)
# The path to the grid directory. Defaults to the test grid directory.
parser.add_argument(
"-grid_dir",
"--grid_dir",
type=str,
required=False,
default=test_grid_dir,
)
# The target metallicity. The code function will find the closest
# metallicity and report it back. The rationale behind this is that this
# code can easily be adapted to explore other grids.
parser.add_argument("-Z", "--Z", type=float, required=False, default=0.01)
# Flag whether to show the figure. Figure is saved in current
# directory using "spectra_age_{grid_name}"
parser.add_argument(
"-show", "--show", action=argparse.BooleanOptionalAction
)
# Flag whether to save the figure.
parser.add_argument(
"-save", "--save", action=argparse.BooleanOptionalAction
)
# Flag whether to analyse all grids in the provided directory.
parser.add_argument("-all", "--all", action=argparse.BooleanOptionalAction)
# Get dictionary of arguments
args = parser.parse_args()
# If all grids are requested get a list of the grids in the grid_dir
# directory.
if args.all:
grid_filenames = list(
map(os.path.basename, glob.glob(args.grid_dir + "*.hdf5"))
)
print(grid_filenames)
# Remove extension
grid_names = list(
map(lambda x: ".".join(x.split(".")[:-1]), grid_filenames)
)
print(grid_names)
# Else use the provided grid name
else:
grid_names = [args.grid_name]
# loop over all grid_names
for grid_name in grid_names:
print(grid_name)
# Initialise grid
grid = Grid(grid_name, grid_dir=args.grid_dir)
# Create figure
fig, ax = plot_spectra_age(grid, args.Z)
# show figure if requested
if args.show:
plt.show()
# save figure if requested
if args.save:
fig.savefig(f"spectra_age_{grid_name}.pdf")
Total running time of the script: (0 minutes 0.513 seconds)